Student Hospitalist Scholar Grant: Derivation And Validation Of A COPD Readmission Risk Prediction Tool
Maximilian Hemmrich
Mentor: Valerie Press, MD, MPH
The goal of this project was to develop a machine learning tool to predict the 90-day readmission risk of patients admitted for COPD exacerbations. Prior work by our team has shown that machine learning techniques significantly increase the validity of predictions, and by only utilizing data readily available from electronic health records (EHRs), this tool could be used in real-time. We also wanted to compare our tool to the PEARL (COPD-specific, 90-day prediction horizon) and HOSPITAL (disease agnostic, 30-day prediction horizon) scores, which are two risk-stratification tools commonly cited in the literature.
In order to train our model, we retrieved EHR encounter data (patient demographics, medications administered, vital signs, laboratory values, etc.) for all admissions at the University of Chicago Medical Center (UCMC) that met our inclusion criteria:
- Age ≥ 40
- Treated with nebulized medications and steroids
- Primary diagnosis of COPD
OR
Primary diagnosis of acute respiratory failure with secondary diagnosis of COPD - Survived encounter
- Unscheduled readmission within 90 days or no readmission within 90 days
This left us with 1,331 patients in our final dataset, of which 306 were readmitted within 90 days. This dataset was then split into two groups: 60% of the data was used as a derivation set, which was used to train the model, and the remaining 40% was used as a validation set, which was used exclusively to assess model performance. We also utilized 10-fold cross-validation during the model derivation phase to improve the model’s generalizability to other datasets.
Using this derivation set, we trained our random forest model in R. Random forests are an ensemble of hundreds, or even thousands, of individual decision trees that each independently predict an outcome for a given input. The prediction of the random forest model is simply the average of the predictions of the individual decision trees. We opted to use a random forest model as previous publications by our team comparing various algorithms in clinical settings showed that random forests outperformed all other algorithms in the study.
Once the model had been trained, we were able to look at the most important predictor variables used by our model:
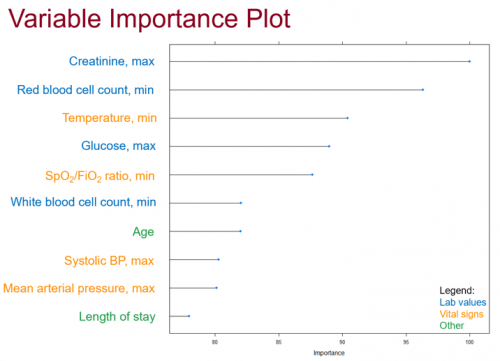
Other predictors that fell outside of the top-10 included whether the patient was admitted during the winter months or not, the results of respiratory cultures, and the patient’s maximum respiratory rate during the encounter.
Our random forest model performed very well in our data set, achieving an area under the receiver-operating characteristic curve (AUROC) of 0.78 and 0.72 in the derivation and validation sets, respectively. These values compared favorably to the published AUCs of both the PEARL and HOSPITAL scores:
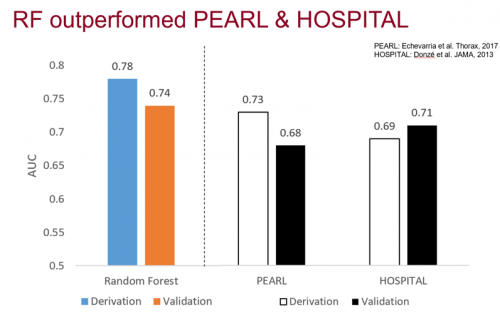
One thing to note is that the AUCs for these models were obtained using different datasets. The PEARL and HOSPITAL scores utilize predictor variables not found in our EHR, preventing us from calculating them with our own dataset at this time.
These results are very promising, although more work needs to go into validating them. This was a single-center study, which could limit generalizability to other medical centers. We also only had access to UCMC readmissions, so patients readmitted elsewhere were not classified correctly in our dataset. Finally, we did not have access to some COPD readmission risk factors commonly cited in the literature, including smoking history, spirometry data, and previous hospitalizations.
Still, there are steps we will take in the future to further improve the model. We plan on obtaining newer EHR data, as more recent versions of the Epic EHR used at UCMC will allow us to access the commonly cited COPD predictors listed above. We also plan on utilizing chart reviews (which we already started this summer), rather than ICD-9 diagnosis codes, to improve the detection of COPD patients and thus increase the usefulness of the model. Lastly, we will calculate the missing PEARL/HOSPITAL predictors in our own dataset so that we can directly compare model performance.
Despite the aforementioned limitations, we were still able to derive and internally validate a random forest machine learning model that can effectively predict whether a COPD patient will be readmitted. The ability to run the mode in real-time using EHR data means it could be inserted into an EHR module for automated risk prediction during the index admission. This would help physicians provide personalized treatment focused on reducing COPD readmissions, improving the quality of care of the patients and reducing medical expenditures.




























































